Note
Go to the end to download the full example code.
Constrained Metamorphosis - simulated cancer growth
This toy example was build to simulate a cancer growth in a brain. The gray scott texture as been used to add intricate patterns to the brain background. This particular example is a very hard registration problem, this example shows how constrained metamorphosis can solve it.
Here we will use the constrained metamorphosis to register two images. The constrained metamorphosis is a metamorphosis that can take into account some prior information to guide the registration. In this example we will use two masks and a prior field to guide the registration:
\(M_t\) : A mask that will control the amount of deformation vs photometric changes.
\(Q_t\) : A mask that will guide the deformation to match a precomputed vector field.
\(w_t\) : A field that will be used to guide the deformation.
The metamorphosis model is defined as follows: Let the image evolution be
The Hamiltonian is defined as
and the deduced geodesic equations are
Import the necessary packages
import matplotlib.pyplot as plt
try:
import sys, os
# add the parent directory to the path
base_path = os.path.join(os.path.dirname(os.path.abspath(__file__)),'..')
sys.path.insert(0,base_path)
import __init__
except NameError:
pass
from demeter.constants import *
import torch
import kornia.filters as flt
# %reload_ext autoreload
# %autoreload 2
import demeter.utils.reproducing_kernels as rk
import demeter.metamorphosis as mt
import demeter.utils.torchbox as tb
device = 'cpu'
if torch.cuda.is_available():
device = 'cuda:0'
print(f"Used device: {device}")
size = (300,300)
location = os.getcwd()
if 'runner' in location:
location = os.path.dirname(os.path.dirname(location))
EXPL_SAVE_FOLDER = os.path.join(location,"saved_optim/")
Used device: cpu
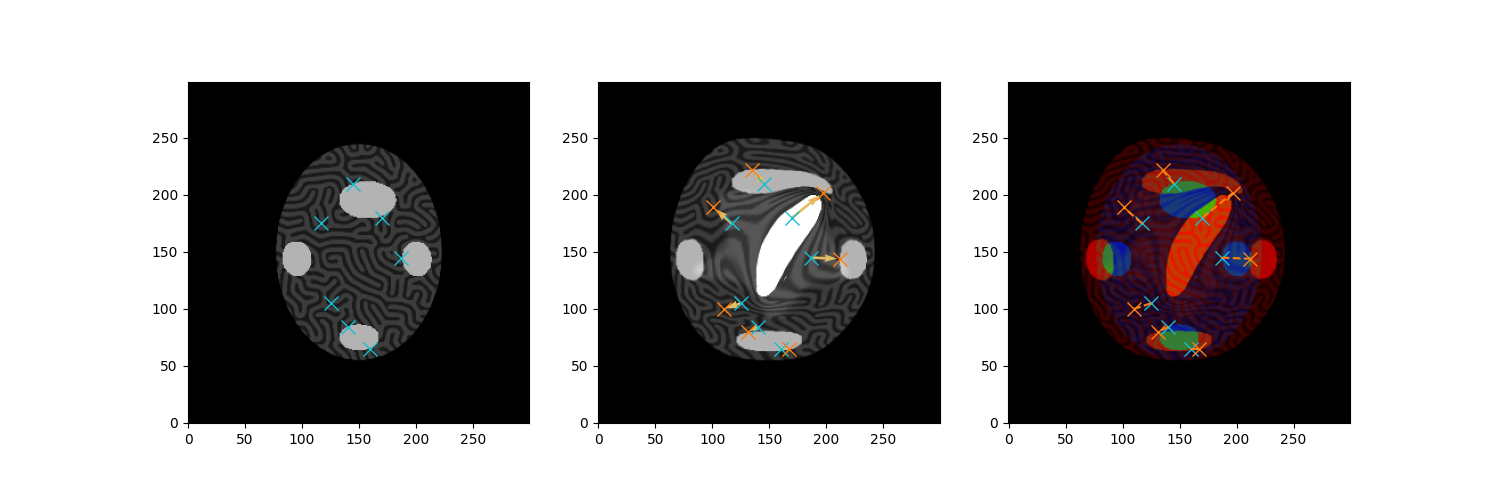
Open and visualise images before registration
source_name,target_name = 'teGS_mbs_S','teGS_mbs_T'
S = tb.reg_open(source_name,size = size).to(device)
T = tb.reg_open(target_name,size = size).to(device)
forDice_source = tb.reg_open('te_s_v_seg',size=size)
forDice_target = tb.reg_open('teGS_mbs_segTdice',size=size)
seg_necrosis = tb.reg_open('teGS_mbs_segNec',size=size).to(device)
seg_oedeme = tb.reg_open('te_o_seg',size=size)
# Put some landmarks on the images to assess registration quality
source_landmarks = torch.Tensor([
[int(187),int(145)],
[int(160),int(65)],
[int(140),int(84)],
[int(145),int(210)],
[int(170),int(180)],
[int(125),int(105)],
[int(117),int(175)]
])
target_landmarks = torch.Tensor([
[int(212),int(144)], # ok
[int(167),int(65)],
[int(131),int(80)], # ok
[int(135),int(222)], # ok
[int(197),int(202)], # ok
[int(110),int(100)], # ok
[int(101),int(189)] # ok
])
id_grid = tb.make_regular_grid(size)
col1 = 'C1'
col2 = 'C9'
fig,ax = plt.subplots(1,3,figsize=(15,5))
ax[0].imshow(S[0,0].cpu(),**DLT_KW_IMAGE)
ax[0].plot(source_landmarks[:,0],source_landmarks[:,1],'x',markersize=10,c=col2)
ax[1].imshow(T[0,0].cpu(),**DLT_KW_IMAGE)
ax[1].plot(source_landmarks[:,0],source_landmarks[:,1],'x',markersize=10,c=col2)
ax[1].plot(target_landmarks[:,0],target_landmarks[:,1],'x',markersize=10,c=col1)
ax[1].quiver(source_landmarks[:,0],source_landmarks[:,1],
target_landmarks[:,0]-source_landmarks[:,0],
target_landmarks[:,1]-source_landmarks[:,1],
color= GRIDDEF_YELLOW)
ax[2].imshow(tb.imCmp(T,S,'seg'),origin='lower')
ax[2].plot(source_landmarks[:,0],source_landmarks[:,1],'x',markersize=10,c=col2)
ax[2].plot(target_landmarks[:,0],target_landmarks[:,1],'x',markersize=10,c=col1)
for i in range(source_landmarks.shape[0]):
ax[2].plot([source_landmarks[i,0],target_landmarks[i,0]],
[source_landmarks[i,1],target_landmarks[i,1]],'--',c=col1)
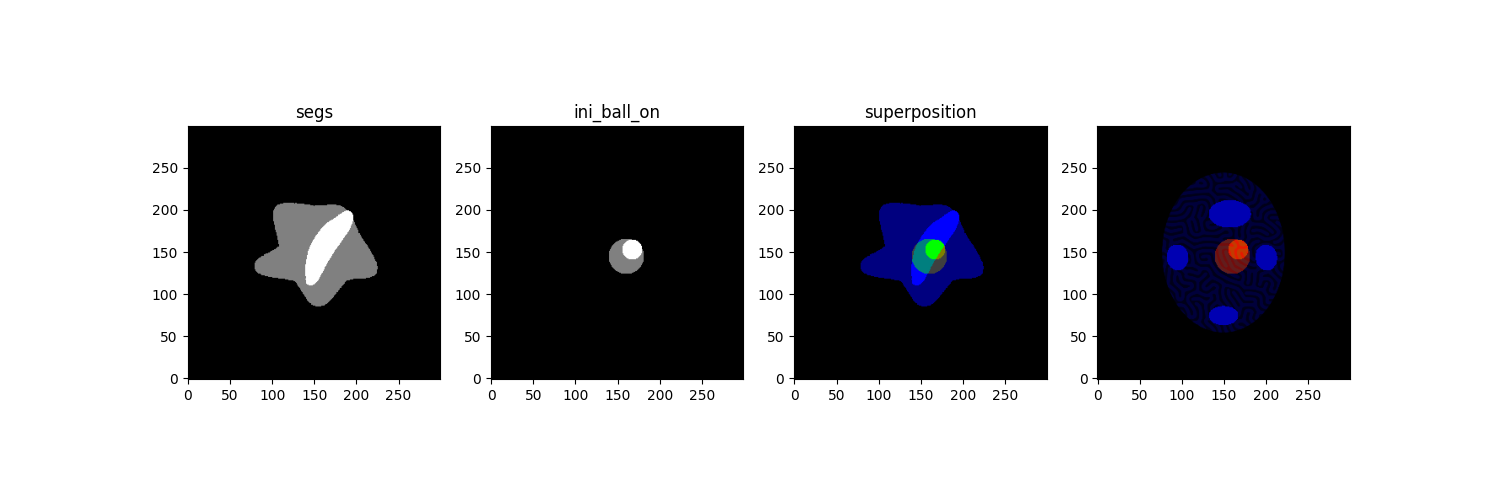
Build masks for constrained metamorphosis. Constrained Metamorphosis can take tree elements as priors: - Residual_mask : a temporal mask with at each pixel a value between 0 and 1
controlling the amount of deformation vs photometric changes.
- orienting_field :a prior precomputed vector field that our deformation
will try to match.
- orienting_maska mask that will be used to weight the orienting field.
for example, if we want to deform only a part, the rest we can indicate it with this mask.
First we need to set source and target masks.
print("`\n==== Building temporal masks ====")
val_o,val_n = .5,1
segs = torch.zeros(seg_necrosis.shape)
segs[seg_oedeme > 0] = val_o
segs[seg_necrosis > 0] = val_n
# plt.imshow(segs[0,0],vmin=0,vmax=1,cmap='gray')
# make source image
center_o = (160,145)
center_n = (167,153)
ini_ball_n,_ = tb.make_ball_at_shape_center(
seg_necrosis,verbose=True,force_r=12,force_center=center_n
)
ini_ball_o,_ = tb.make_ball_at_shape_center(
seg_necrosis,verbose=True,force_r=21,force_center=center_o
)
ini_ball_on = torch.zeros(ini_ball_o.shape)
ini_ball_on[ini_ball_o > 0] = val_o
ini_ball_on[ini_ball_n > 0] = val_n
# segs = torch.ones_like(segs) * .5
# ini_ball_on = torch.ones_like(ini_ball_on) * .5
fig, ax = plt.subplots(1,4,figsize=(15,5))
ax[0].imshow(segs[0,0],vmin=0,vmax=1,cmap='gray',origin='lower')
ax[0].set_title('segs')
ax[1].imshow(ini_ball_on[0,0],vmin=0,vmax=1,cmap='gray',origin='lower')
ax[1].set_title('ini_ball_on')
ax[2].imshow(tb.imCmp(ini_ball_on,segs,'seg'),origin='lower')
ax[2].set_title('superposition')
ax[3].imshow(tb.imCmp(ini_ball_on,S,'seg'),origin='lower')
plt.show()
`
==== Building temporal masks ====
centre = (167, 153), r = 12 and the seg and ball have 364 pixels overlapping
centre = (160, 145), r = 21 and the seg and ball have 1001 pixels overlapping
fix other constants
## Build temporal masks
sigma = [(10,10),(15,15)]
# sigma = [(10,10)]
kernelOp = rk.Multi_scale_GaussianRKHS(sigma,normalized=True)
# rk.plot_kernel_on_image(kernelOp,subdiv=10,image=T.cpu())
plt.show()
print(kernelOp)
dx_convention = 'pixel'
n_steps= 10
Multi_scale_GaussianRKHS(
sigma :[(10, 10), (15, 15)],
kernel size :(1, 91, 91)
)
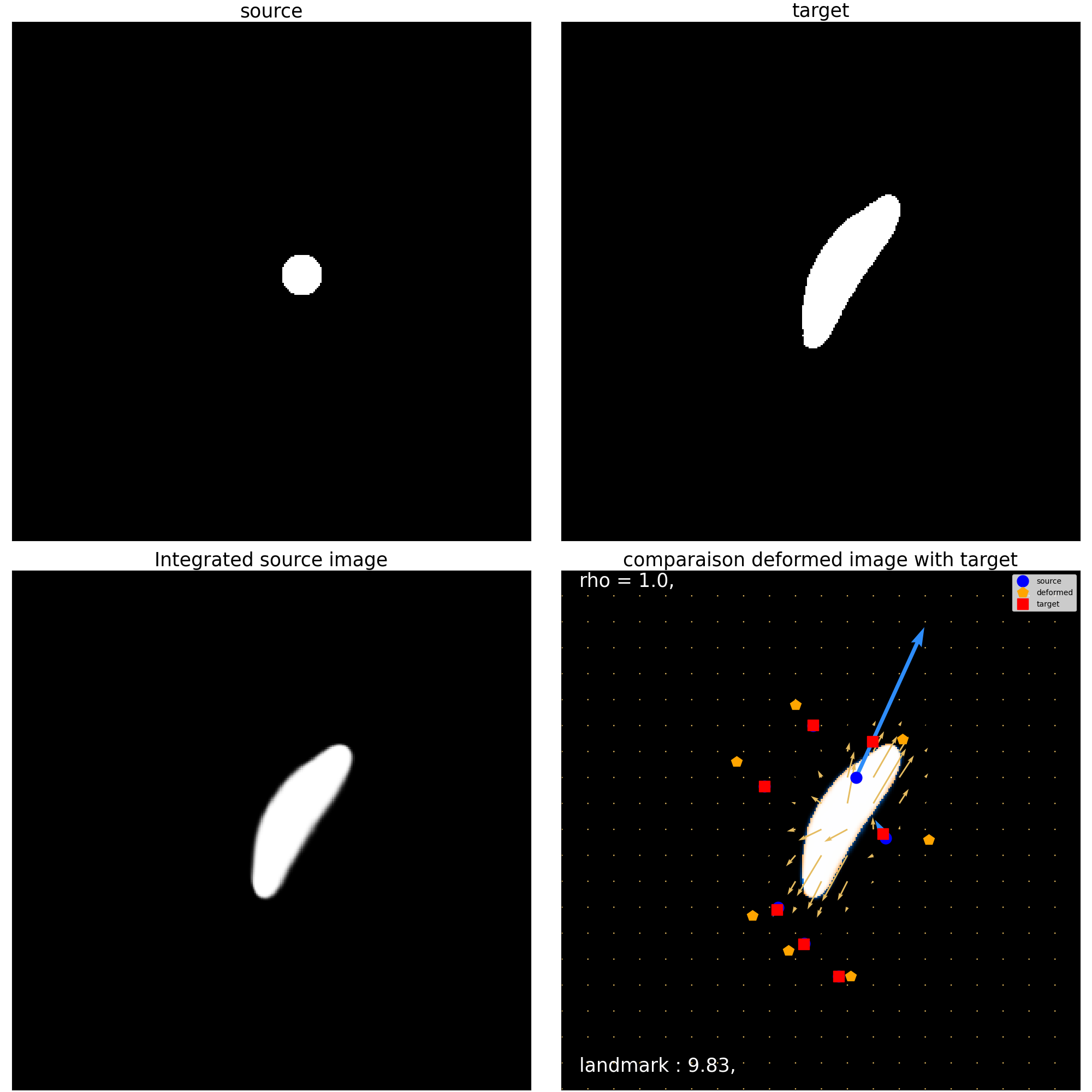
print(">>>> Mask for orienting field <<<<")
recompute = False
if recompute:
momentum_ini = 0
mr_mask_orienting = mt.lddmm(ini_ball_n.to(device),seg_necrosis,momentum_ini,
kernelOperator=kernelOp,cost_cst=1e-5,integration_steps=n_steps,
n_iter=10,grad_coef=1,
optimizer_method='LBFGS_torch',
dx_convention=dx_convention,)
mr_mask_orienting.save(f"mask_tE_gs_CM_{dx_convention}_n_step{n_steps}_orienting")
else:
file = "2D_11_02_2025_mask_tE_gs_CM_pixel_n_step10_orienting_000.pk1"
mr_mask_orienting = mt.load_optimize_geodesicShooting(file,
path =EXPL_SAVE_FOLDER
)
mr_mask_orienting.compute_landmark_dist(source_landmarks,target_landmarks)
mr_mask_orienting.plot_imgCmp()
plt.show()
# # #%%
# mr_mask_orienting.plot_deform()
# mr_mask_orienting.mp.plot()
# plt.show()
>>>> Mask for orienting field <<<<
DT: None
New optimiser loaded (2D_11_02_2025_mask_tE_gs_CM_pixel_n_step10_orienting_000.pk1) :
Metamorphosis_Shooting(cost_parameters : {,
rho =1.0,
lambda =1e-05
},
geodesic integrator : Metamorphosis_integrator(
(kernelOperator): Multi_scale_GaussianRKHS(
sigma :[(10, 10), (15, 15)],
kernel size :(1, 91, 91)
)
)
integration method : _step_full_semiLagrangian
optimisation method : LBFGS_torch
# geodesic steps =10
)
round False
round False
Landmarks:
Before : 11.928571701049805
After : 9.826631546020508
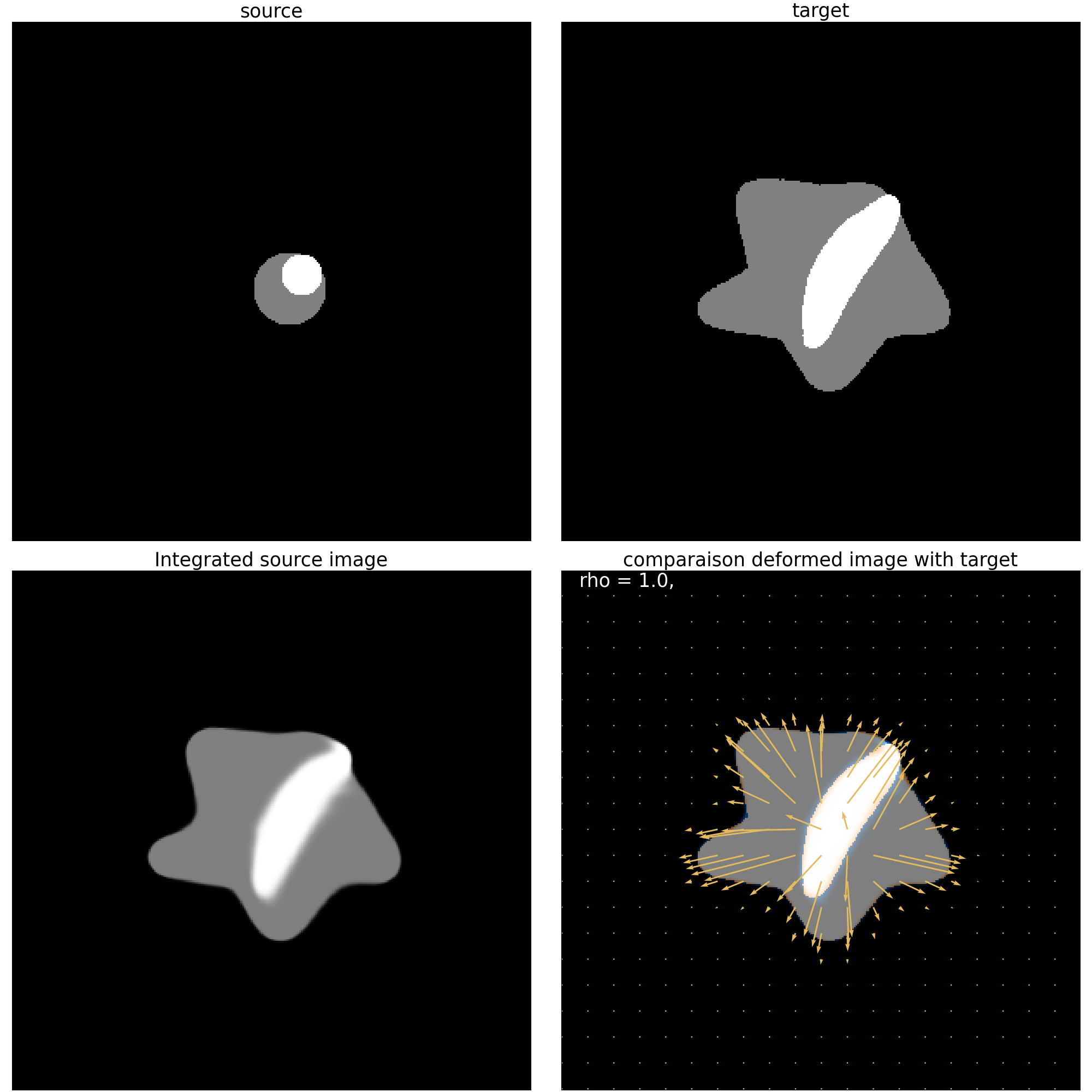
print(">>>> Mask for residuals field <<<<")
if recompute:
momentum_ini = 0
mr_mask_residuals = mt.lddmm(ini_ball_on.to(device),segs,momentum_ini,
kernelOperator=kernelOp,cost_cst=1e-5,integration_steps=n_steps,
n_iter=15,grad_coef=1,
optimizer_method='LBFGS_torch',
dx_convention=dx_convention,)
mr_mask_residuals.save(f"mask_tE_gs_CM_{dx_convention}_n_step{n_steps}_residuals")
else:
file = "2D_11_02_2025_mask_tE_gs_CM_pixel_n_step10_residuals_000.pk1"
mr_mask_residuals = mt.load_optimize_geodesicShooting(file, path =EXPL_SAVE_FOLDER)
mr_mask_residuals.plot_imgCmp()
plt.show()
>>>> Mask for residuals field <<<<
DT: None
New optimiser loaded (2D_11_02_2025_mask_tE_gs_CM_pixel_n_step10_residuals_000.pk1) :
Metamorphosis_Shooting(cost_parameters : {,
rho =1.0,
lambda =1e-05
},
geodesic integrator : Metamorphosis_integrator(
(kernelOperator): Multi_scale_GaussianRKHS(
sigma :[(10, 10), (15, 15)],
kernel size :(1, 91, 91)
)
)
integration method : _step_full_semiLagrangian
optimisation method : LBFGS_torch
# geodesic steps =10
)
mr_mask_residuals.mp.plot() plt.show()
The exercise here is too model the mask and tweak the masks to make the expected registration. Keep in mind that masks should be between 0 and 1
residuals_mask = mr_mask_residuals.mp.image_stock.clone()
residuals_mask = 1 - residuals_mask
orienting_field = -mr_mask_orienting.mp.field_stock.clone() / n_steps
norm_w_2 = (orienting_field ** 2).sum(dim= -1).sqrt()
norm_w_2 = norm_w_2/norm_w_2.max()
orienting_mask = norm_w_2.clone()[:,None]
o_max = 0.02
orienting_mask[orienting_mask > o_max] = o_max
sig = 5 # blur the mask to avoid sharp transitions
residuals_mask = flt.gaussian_blur2d(residuals_mask,(int(6*sig)+1,int(6*sig)+1),(sig,sig))
# sig = 5
orienting_mask = flt.gaussian_blur2d(orienting_mask,(int(6*sig)+1,int(6*sig)+1),(sig,sig))
L = [0,2,8,-1]
fig,ax = plt.subplots(2,len(L),figsize=(len(L)*5,10))
ax[0,0].set_title('orienting mask')
ax[1,0].set_title('residuals mask')
for i,ll in enumerate(L):
ax[0,i].imshow(orienting_mask[ll,0].cpu(),cmap='gray',origin = "lower",
# vmin=0, vmax = 1,
)
tb.quiver_plot(orienting_mask[ll,0][...,None].cpu() * orienting_field[ll][None].cpu(),
ax[0,i],
step = 10,color='C3',dx_convention=dx_convention)
ax[1,i].imshow(residuals_mask[ll,0].cpu(),cmap='gray',vmin=0, vmax = 1,origin = "lower")
plt.show()
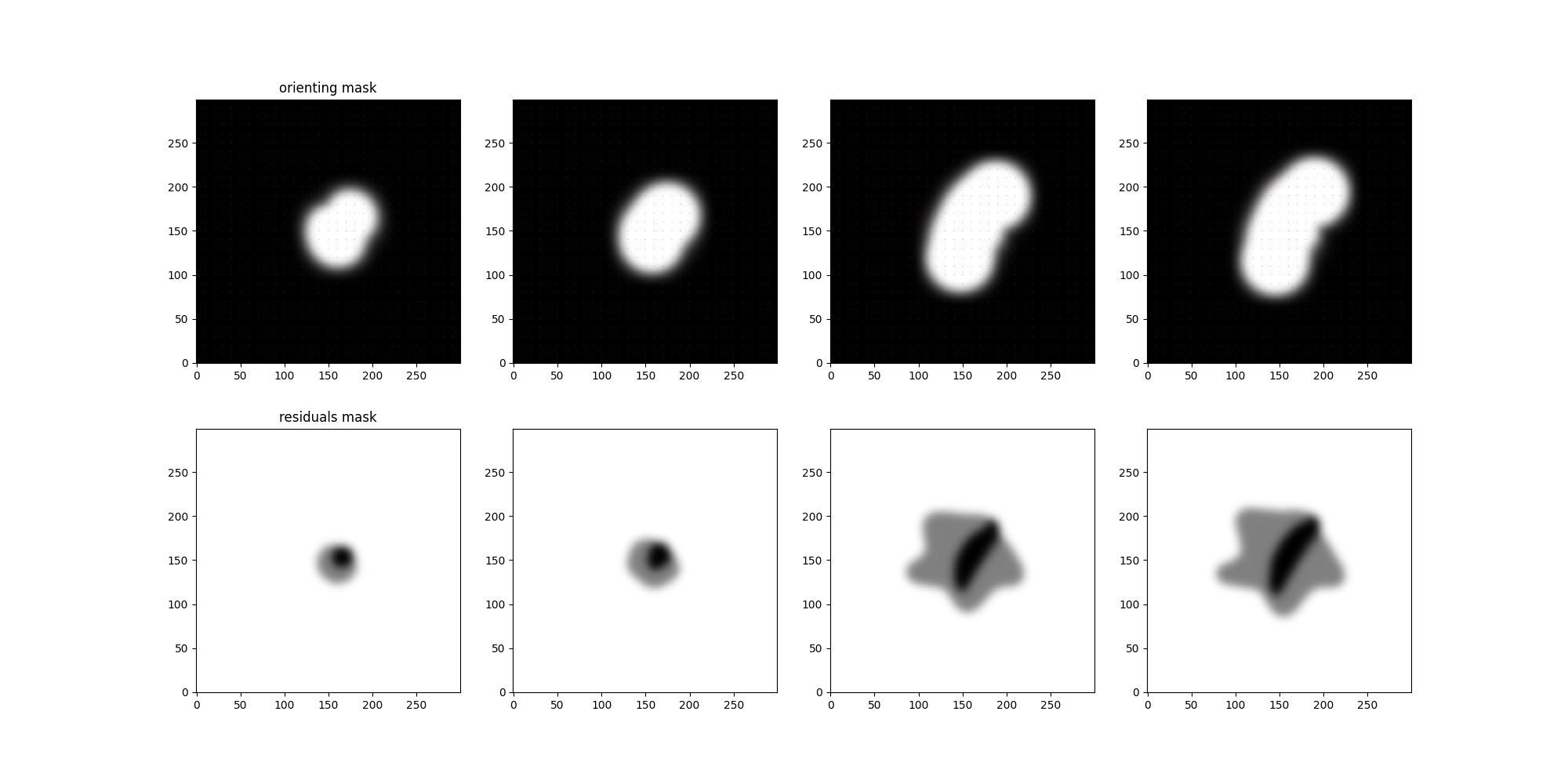
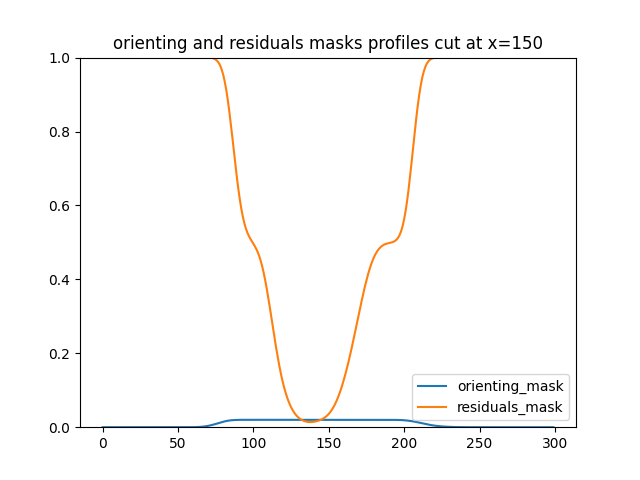
fig1,ax1 = plt.subplots(1,1)
ax1.plot(orienting_mask[-1,0,:,150].cpu(),label="orienting_mask")
ax1.plot(residuals_mask[-1,0,:,150].cpu(),label="residuals_mask")
ax1.set_ylim(0,1)
ax1.legend()
plt.title('orienting and residuals masks profiles cut at x=150')
# l = orienting_mask.shape[0]
# L = range(l)
# fig,ax = plt.subplots(l,1,figsize=(10,l*10))
plt.show()
sigma = [(5,5),(10,10),(15,15)]
# sigma = [(10,10)]
kernelOp = rk.Multi_scale_GaussianRKHS(sigma,normalized=False)
# kernelOp.kernel = kernelOp.kernel / kernelOp.kernel.max()
kernelOp.plot()
plt.show()
print(kernelOp)
![kernel sigma: (10, 10), Gaussian Kernel $\sigma$=[(5, 5), (10, 10), (15, 15)], kernel sigma: (5, 5)](../../_images/sphx_glr_toyExample_grayScott_constrainedMetamorphosis_007.png)
sigma is not a float, I suspect that the kernel is not purly gaussian.
/home/runner/work/Demeter_metamorphosis/Demeter_metamorphosis/src/demeter/utils/reproducing_kernels.py:307: UserWarning: No artists with labels found to put in legend. Note that artists whose label start with an underscore are ignored when legend() is called with no argument.
ax.legend()
sigma is not a float, I suspect that the kernel is not purly gaussian.
Multi_scale_GaussianRKHS(
sigma :[(5, 5), (10, 10), (15, 15)],
kernel size :(1, 91, 91)
)
orienting_mask = None you can also load the optimisation object from a file file = “2D_25_01_2025_TEST_toyExample_grayScott_CM_square_n_step20_000.pk1” mr = mt.load_optimize_geodesicShooting(file)
print("\n==== Constrained Metamorphosis ====")
momentum_ini = 0
ic.disable()
mr_cm = mt.constrained_metamorphosis(S,T,momentum_ini,
orienting_mask,
orienting_field,
residuals_mask,
kernelOperator=kernelOp,
cost_cst=1e-10,
grad_coef=.1,
n_iter=20,
dx_convention=dx_convention,
# optimizer_method='adadelta',
)
if recompute:
mr_cm.compute_landmark_dist(source_landmarks,target_landmarks)
mr_cm.plot_cost()
plt.show()
mr_cm.save(f"toyExample_grayScott_CM_{dx_convention}_n_step{n_steps}")
else:
mr_cm = mt.load_optimize_geodesicShooting("2D_11_02_2025_toyExample_grayScott_CM_pixel_n_step10_000.pk1",
path =EXPL_SAVE_FOLDER
)
==== Constrained Metamorphosis ====
oriented
Weighted
Progress: [#---------] 10.00% (Ssd : ,187.2694).
Progress: [##--------] 15.00% (Ssd : , 99.0902).
Progress: [##--------] 20.00% (Ssd : , 67.8786).
Progress: [##--------] 25.00% (Ssd : , 55.3011).
Progress: [###-------] 30.00% (Ssd : , 48.4873).
Progress: [####------] 35.00% (Ssd : , 45.3169).
Progress: [####------] 40.00% (Ssd : , 42.4989).
Progress: [####------] 45.00% (Ssd : , 40.6994).
Progress: [#####-----] 50.00% (Ssd : , 39.1277).
Progress: [######----] 55.00% (Ssd : , 38.0623).
Progress: [######----] 60.00% (Ssd : , 37.1787).
Progress: [######----] 65.00% (Ssd : , 36.2048).
Progress: [#######---] 70.00% (Ssd : , 35.4607).
Progress: [########--] 75.00% (Ssd : , 34.8593).
Progress: [########--] 80.00% (Ssd : , 34.2708).
Progress: [########--] 85.00% (Ssd : , 33.9125).
Progress: [#########-] 90.00% (Ssd : , 33.5998).
Progress: [##########] 95.00% (Ssd : , 33.2848).
Progress: [##########] 100.00% Done...
(Ssd : , 32.9175).
Computation of forward done in 0:01:43s and 0.827cents s
Computation of constrained_metamorphosis done in 0:01:43s and 0.828cents s
DT: None
round False
round False
Landmarks:
Before : 11.928571701049805
After : 5.454163551330566
New optimiser loaded (2D_11_02_2025_toyExample_grayScott_CM_pixel_n_step10_000.pk1) :
ConstrainedMetamorphosis_Shooting(cost_parameters : {,
rho =Weighted,
lambda =1e-10
},
geodesic integrator : ConstrainedMetamorphosis_integrator(Weighted & Oriented)
n_step = 10
Multi_scale_GaussianRKHS(
sigma :[(5, 5), (10, 10), (15, 15)],
kernel size :(1, 91, 91)
)
integration method : step
optimisation method : LBFGS_torch
# geodesic steps =10
)
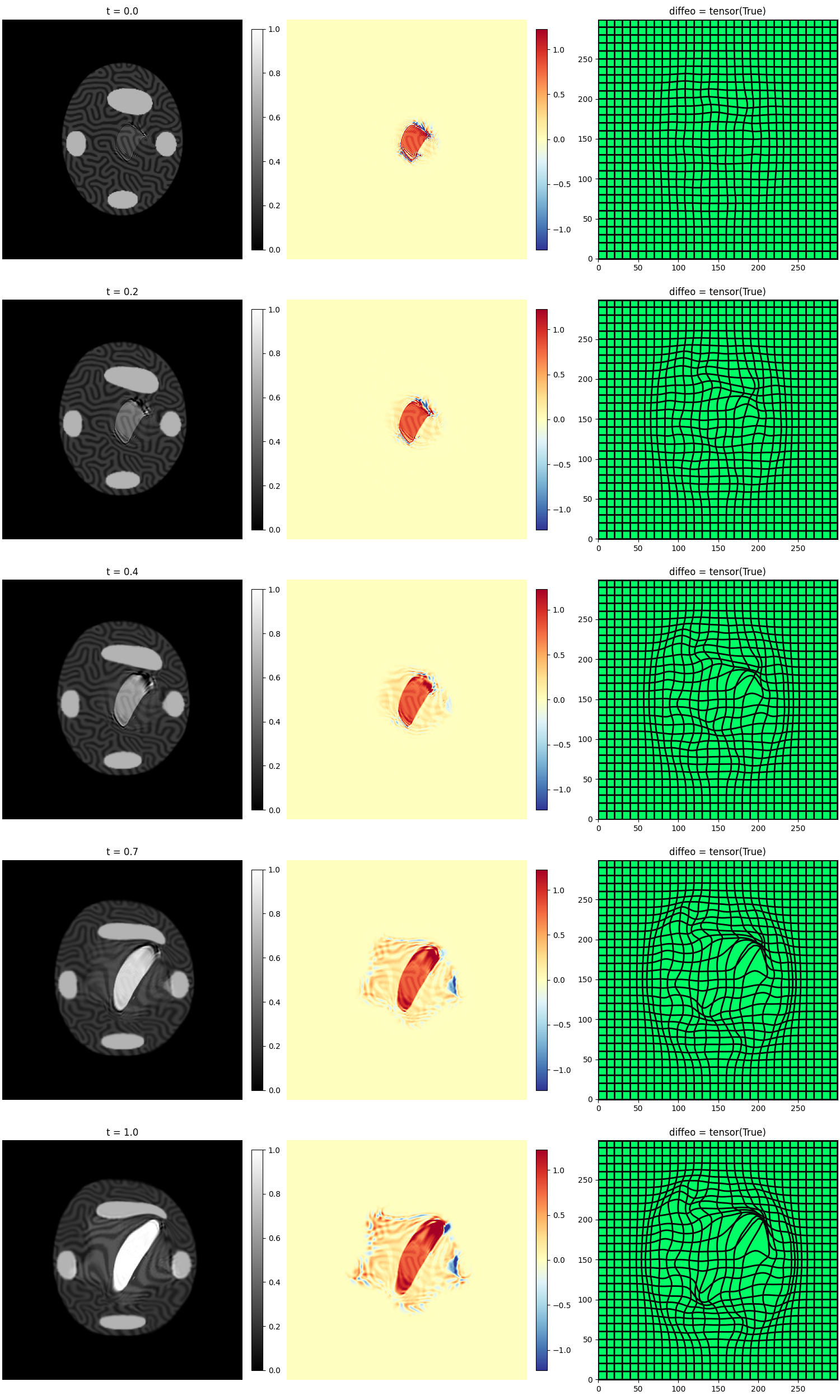
mr_cm.plot_imgCmp()
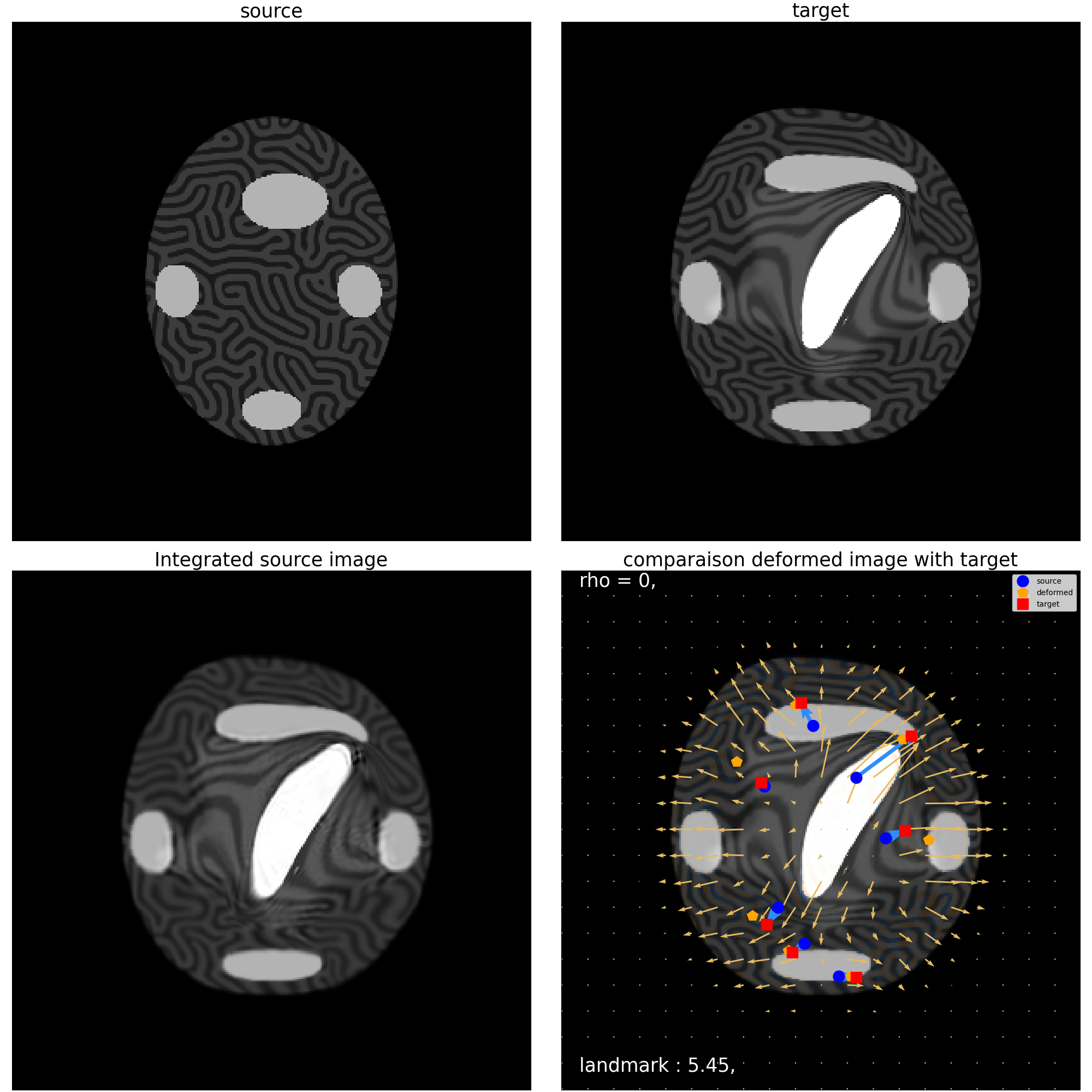
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers). Got range [-0.03132463991641998..1.1656873226165771].
(<Figure size 2000x2000 with 4 Axes>, array([[<Axes: title={'center': 'source'}>,
<Axes: title={'center': 'target'}>],
[<Axes: title={'center': 'Integrated source image'}>,
<Axes: title={'center': 'comparaison deformed image with target'}>]],
dtype=object))
mr_cm.plot_deform()
plt.show()
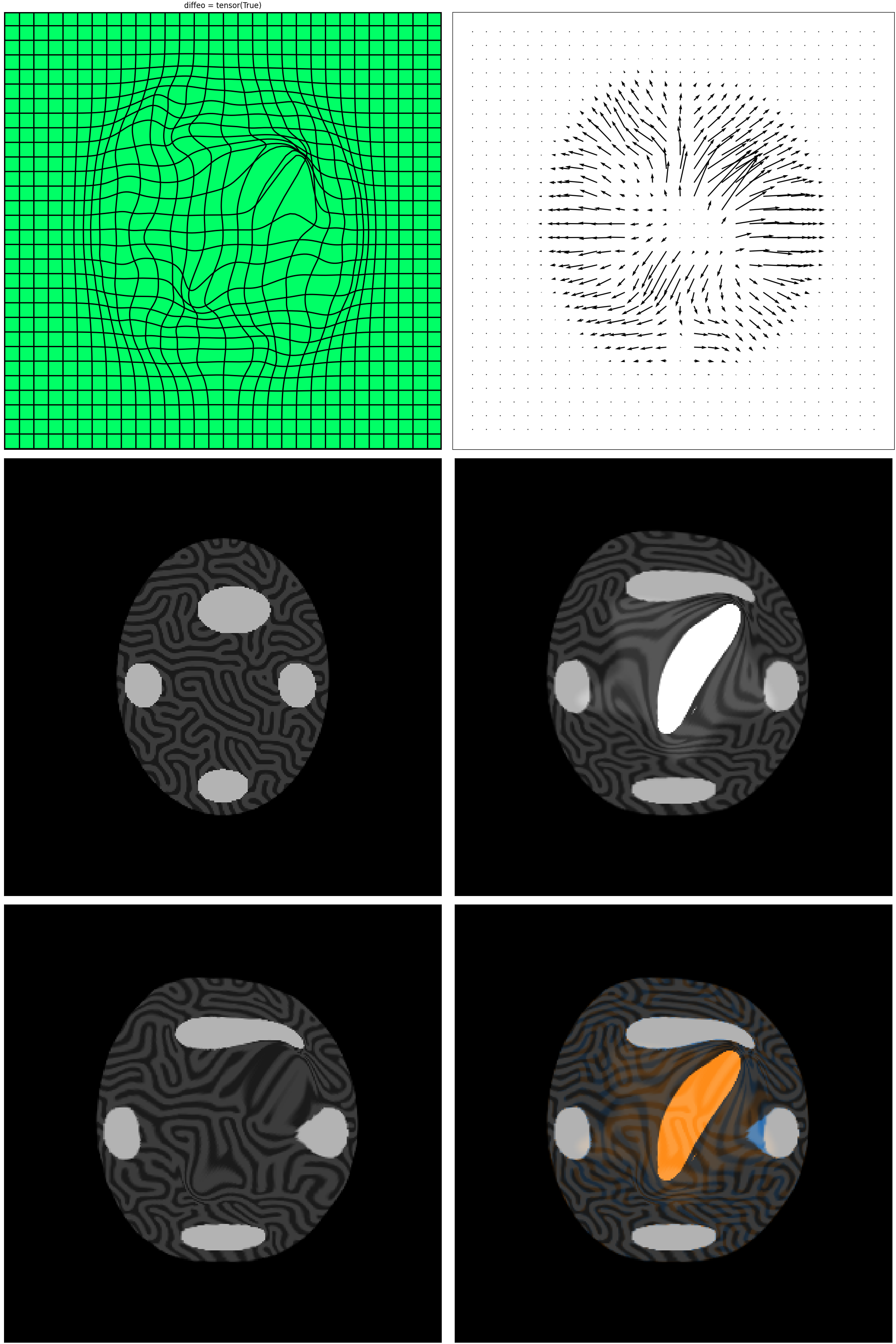
mr_cm.mp.plot()
plt.show()
# mr_cm.save_to_gif("image",f"toyExample_grayScott_CM_{dx_convention}_n_step{n_steps}_image",
# folder='toyExample_grayScott')
mr_cm.save_to_gif("residual",f"toyExample_grayScott_CM_{dx_convention}_n_step{n_steps}_residual",
folder='toyExample_grayScott')
convert -delay 40 -loop 0 /home/runner/work/Demeter_metamorphosis/Demeter_metamorphosis/examples/gifs/toyExample_grayScott/toyExample_grayScott_CM_pixel_n_step10_residual_\d3.png /home/runner/work/Demeter_metamorphosis/Demeter_metamorphosis/examples/gifs/toyExample_grayScott/toyExample_grayScott_CM_pixel_n_step10_residual.gif
Cleaning saved files.
Your gif was successfully saved at : /home/runner/work/Demeter_metamorphosis/Demeter_metamorphosis/examples/gifs/toyExample_grayScott/toyExample_grayScott_CM_pixel_n_step10_residual.gif
('/home/runner/work/Demeter_metamorphosis/Demeter_metamorphosis/examples/gifs/toyExample_grayScott/', 'toyExample_grayScott_CM_pixel_n_step10_residual.gif')
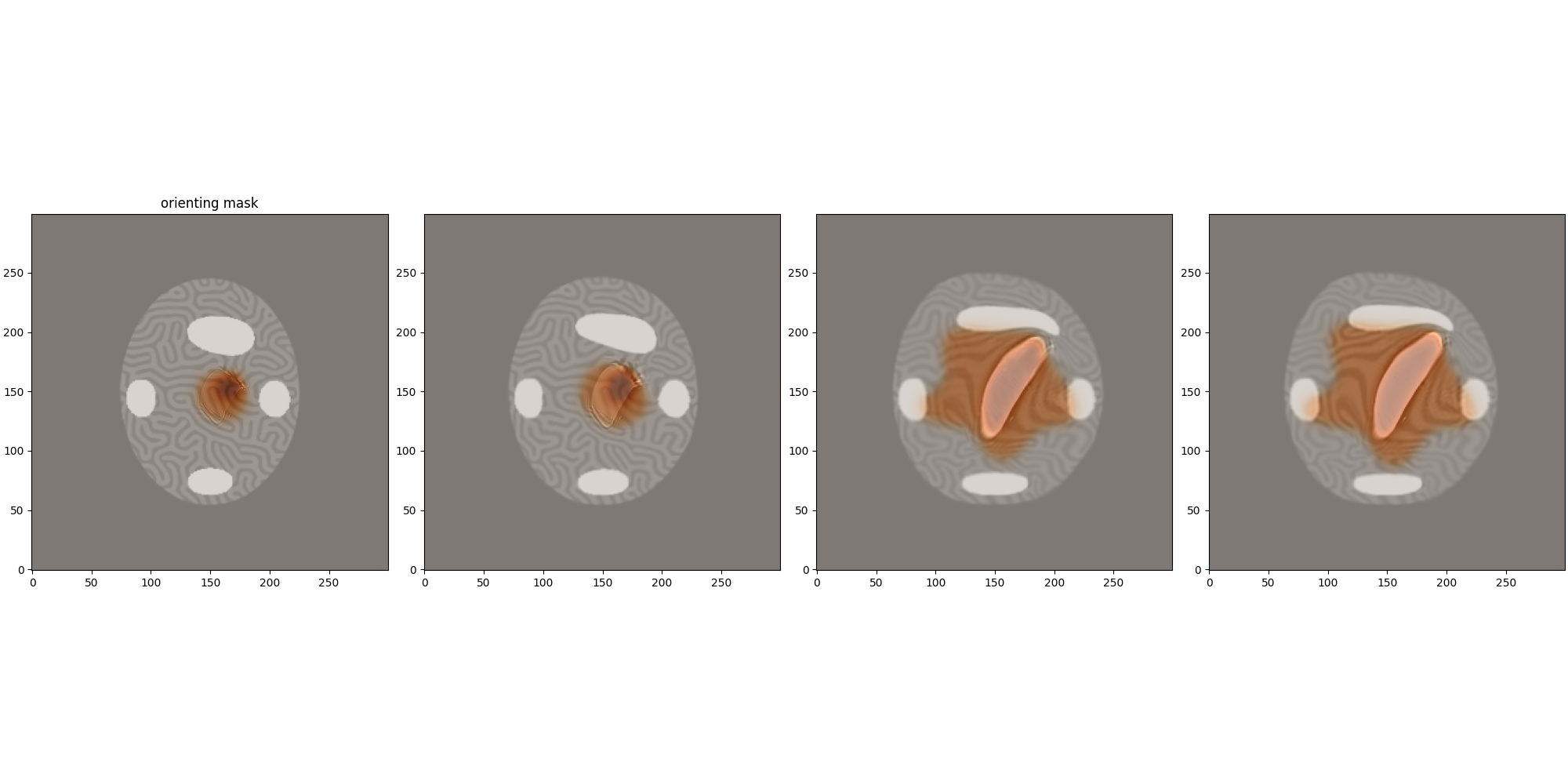
L = [0,2,8,-1]
fig,ax = plt.subplots(1,len(L),figsize=(len(L)*5,10), constrained_layout=True)
ax[0].set_title('orienting mask')
for i,ll in enumerate(L):
ax[i].imshow(mr_cm.mp.image_stock[ll,0].cpu(),cmap='gray',vmin=0, vmax = 1,origin = "lower")
ax[i].imshow(1-residuals_mask[ll,0].cpu(),cmap='Oranges',vmin=0, vmax = 1,origin = "lower",alpha = .5)
# ax[i].imshow(orienting_mask[ll,0].cpu(),cmap='Blues',vmin=0, vmax = 1,origin = "lower",alpha = .5)
plt.show()
Total running time of the script: (1 minutes 50.591 seconds)